The AuraSeq-Comprehensive Panel is a highly multiplexed assay based on a next-generation sequencing platform. AuraSeq-Comprehensive identifies clinically relevant alterations in 500 cancer-related genes, including base pair substitutions, small insertions and deletions in thousands of mutational hotspots, as well as copy number variations, intergenic and intragenic fusions, and genomic signatures, allowing physicians to expand treatment options to include most FDA-approved targeted therapies.
The AuraSeq-Comprehensive Panel is a highly multiplexed assay based on a next-generation sequencing platform. AuraSeq-Comprehensive identifies clinically relevant alterations in 500 cancer-related genes, including base pair substitutions, small insertions and deletions in thousands of mutational hotspots, as well as copy number variations, intergenic and intragenic fusions, and genomic signatures, allowing physicians to expand treatment options to include most FDA-approved targeted therapies.
Covers over 500 unique genes, to encompass 1.4 Mb of genome, and 1300 gene fusions
Detects single-gene biomarkers such as SNVs, indels, CNVs, fusions, and splice variants (for Molecularly Targeted Therapies)
Detects homologous recombination deficiency (HRD) signatures by assessing variants in 42 HRD genes and genomic instability with sample-level loss of heterozygosity (LOH) in key HRD genes, including BRCA1 and BRCA2 (for PARP inhibitors)
Renders complex multi-gene biomarkers for mutational signatures, including tumor mutational burden (TMB) and microsatellite instability (MSI) (for Immune Checkpoint Inhibitors)
AuraSeq-Comprehensive upholds low QNS rates with high sequencing success rates (>98%) with as little as 2-20 ng of DNA/RNA from FFPE samples1
Highly automated workflow and streamlined bioinformatics analysis pipeline optimized for delivering results with a short turnaround time (TAT = 7-12 days)
TEST DESCRIPTION
The AuraSeq-Comprehensive Panel utilizes next-generation sequencing (NGS) technology to detect cancer-related genomic alterations such as single nucleotide variants (SNVs), small insertions and deletions (Indels), copy number variations (CNVs), and gene fusions; as well as genomic signatures such as tumor mutational burden (TMB), microsatellite instability (MSI), and homologous recombination (HR) repair gene alterations and genomic instability due to HR deficiency (HRD).
This test is intended to detect genomic alterations in several metastatic or advanced cancers, including:
-
Non-small cell lung cancer (NSCLC) (e.g.: EGFR, BRAF, ERBB2, KRAS, MET (METex14 and METamp), ALK-fusions, ROS1-fusions, RET-fusions). These genomic biomarkers are recommended by national guidelines.
-
Colorectal cancer (CRC) (e.g.: MSI, KRAS, NRAS, BRAF, ROS1-fusions, NTRK1-fusions, NTRK3-fusions). Most of these biomarkers are recommended by national guidelines.
-
Spitzoid melanoma (e.g.: TMB, NTRK1-fusions)
-
Gliomas (e.g.: IDH1, IDH2, EGFRamp, EGFRvIII isoform, 1p/19q co-deletion, NTRK1-fusions, FGFR1-fusions, FGFR3-fusions)
-
Bladder cancer (FGFR3-fusions)
-
Breast cancer (e.g.: BRCA1, BRCA2, HR genes, NTRK3-fusions, FGFR1-fusions, FGFR2-fusions, FGFR3-fusions)
-
Ovarian cancer (e.g.: HRD status, BRCA1, BRCA2, HR genes, NTRK3-fusions, FGFR1-fusions, FGFR2-fusions, and FGFR3-fusions)
-
Prostate cancer (e.g.: HRD status, ERG-fusions, ETV1-fusions, ETV4-fusions, FGFR1-fusions)
-
Pancreatic cancer (e.g.: HRD status, KRAS, SMAD4, NTRK1/2/3-fusions, ROS1-fusions)
-
Soft tissue sarcomas (e.g.: NTRK1-fusions, NTRK3 -fusions)
-
Any other advanced carcinoma which has failed to respond to conventional chemotherapy or is about to start tyrosine kinase inhibitors (TKIs), an immune checkpoint inhibitor (ICI) or a poly (ADP ribose) polymerase (PARP) inhibitor regime.
The detection of cancer-related SNVs, small Indels, and structural variants (SVs), including CNVs and gene fusions using NGS technology in solid tumors, has shown great clinical utility by providing informative genomic data to assist health care providers in making a diagnosis of cancer, estimating prognosis, and making treatment decisions involving molecular targeted drugs.2, 3 As new molecular targeted therapies become available in clinical practice, detection of targetable gene variants as well as genomic signatures is increasingly important in order to drive therapy of choice.
The AuraSeq Impact
Variant Types Detected
Validated for Small Samples - AuraSeq requires minimal material, with cytology smears being acceptable.4, 5
Great Success Rate - With a <2% QNS rate6, lower than other leading NGS-based test providers, including samples with low tumor content (5-10% neoplastic nuclei).7
Actionable Results for Oncology - Provides information on targets of >85 FDA-approved therapies, and clinical trial eligibility.
| TAT | 7-12 days |
| Sample Type |
FFPE; Cytology Smears |
| DNA Input |
2-20 ng |
| RNA Input |
2-20 ng |
| Failure Rate |
5.6%5 |
| No. of Genes |
500+, 1,300 fusions |
| LoD |
4% |
The AuraSeq-Comprehensive test assesses both DNA and RNA for a uniquely comprehensive and highly accurate detection of actionable biomarkers for solid tumor cancers utilizing very limited tissue or cytology smears.
| Genes & Fusions |
> 500 genes + > 1300 fusions |
| DNA | |
| RNA | |
| Hotspots (SNVs + Indels) | |
| CNVs | |
| Fusions | |
| MSI | |
| TMB | |
| HRD | |
| Long Indels |
Hot Spots (SNVs and Small Indels)
| ABL1 | CUL1 | HIST1H2BD | NFE2L2 | RICTOR |
| ABL2 | CYSLTR2 | HIST1H3B | NRAS | RIT1 |
| ACVR1 | DDR2 | HRAS | NSD2 | ROS1 |
| AKT1 | DGCR8 | IDH1 | NT5C2 | RPL10 |
| AKT2 | DROSHA | IDH2 | NTRK1 | SETBP1 |
| AKT3 | E2F1 | IKBKB | NTRK2 | SF3B1 |
| ALK | EGFR | IL6ST | NTRK3 | SIX1 |
| AR | EIF1AX | IL7R | NUP93 | SIX2 |
| ARAF | EPAS1 | IRF4 | PAX5 | SLCO1B3 |
| ATP1A1 | ERBB2 | IRS4 | PCBP1 | SMC1A |
| AURKA | ERBB3 | KDR | PDGFRA | SMO |
| AURKC | ERBB4 | KIT | PDGFRB | SNCAIP |
| AXL | ESR1 | KLF4 | PIK3C2B | SOS1 |
| BCL2 | EZH2 | KLF5 | PIK3CA | SOX2 |
| BCL2L12 | FAM135B | KNSTRN | PIK3CB | SPOP |
| BCL6 | FGF7 | KRAS | PIK3CD | SRC |
| BCR | FGFR1 | MAGOH | PIK3CG | SRSF2 |
| BMP5 | FGFR2 | MAP2K1 | PIK3R2 | STAT3 |
| BRAF | FGFR3 | MAP2K2 | PIM1 | STAT5B |
| BTK | FGFR4 | MAPK1 | PLCG1 | STAT6 |
| CACNA1D | FLT3 | MAX | PPP2R1A | TAF1 |
| CARD11 | FLT4 | MDM4 | PPP6C | TERT |
| CBL | FOXA1 | MECOM | PRKACA | TGFBR1 |
| CCND1 | FOXL2 | MED12 | PTPN11 | TOP1 |
| CCND2 | FOXO1 | MEF2B | PTPRD | TPMT |
| CCND3 | GATA2 | MET | PXDNL | TRRAP |
| CCNE1 | GLI1 | MITF | RAC1 | TSHR |
| CD79B | GNA11 | MPL | RAF1 | U2AF1 |
| CDK4 | GNAQ | MTOR | RARA | USP8 |
| CDK6 | GNAS | MYC | RET | WAS |
| CHD4 | H3F3A | MYCN | RGS7 | XPO1 |
| CSF1R | H3F3B | MYD88 | RHEB | ZNF217 |
| CTNNB1 | HIF1A | MYOD1 | RHOA | ZNF429 |
CNVs
| ABCB1 | CHEK2 | GATA2 | NBN | RHEB |
| ABL1 | CIC | GATA3 | NCOR1 | RICTOR |
| ABL2 | CREBBP | GLI3 | NF1 | RIT1 |
| ABRAXAS1 | CSMD3 | GNA13 | NF2 | RNASEH2A |
| ACVR1B | CTCF | GNAS | NFE2L2 | RNASEH2B |
| ACVR2A | CTLA4 | GPS2 | NOTCH1 | RNF43 |
| ADAMTS12 | CTNND2 | H3F3A | NOTCH2 | ROS1 |
| ADAMTS2 | CUL3 | H3F3B | NOTCH3 | RPA1 |
| AKT1 | CUL4A | HDAC2 | NOTCH4 | RPS6KB1 |
| AKT2 | CUL4B | HDAC9 | NRAS | RPTOR |
| AKT3 | CYLD | HLA-A | NTRK1 | RUNX1 |
| ALK | CYP2C9 | HLA-B | NTRK3 | SDHA |
| AMER1 | DAXX | HNF1A | PALB2 | SDHB |
| APC | DDR1 | IDH2 | PARP1 | SDHD |
| AR | DDR2 | IGF1R | PARP2 | SETBP1 |
| ARAF | DDX3X | IKBKB | PARP3 | SETD2 |
| ARHGAP35 | DICER1 | IL7R | PARP4 | SF3B1 |
| ARID1A | DNMT3A | INPP4B | PBRM1 | SLCO1B3 |
| ARID1B | DOCK3 | JAK1 | PCBP1 | SLX4 |
| ARID2 | DPYD | JAK2 | PDCD1 | SMAD2 |
| ARID5B | DSC1 | JAK3 | PDCD1LG2 | SMAD4 |
| ASXL1 | DSC3 | KDM5C | PDGFRA | SMARCA4 |
| ASXL2 | EGFR | KDM6A | PDGFRB | SMARCB1 |
| ATM | EIF1AX | KDR | PDIA3 | SMC1A |
| ATR | ELF3 | KEAP1 | PGD | SMO |
| ATRX | EMSY | KIT | PHF6 | SOX9 |
| AURKA | ENO1 | KLF5 | PIK3C2B | SPEN |
| AURKC | EP300 | KMT2A | PIK3CA | SPOP |
| AXIN1 | EPCAM | KMT2B | PIK3CB | SRC |
| AXIN2 | EPHA2 | KMT2C | PIK3R1 | STAG2 |
| AXL | ERAP1 | KMT2D | PIK3R2 | STAT3 |
| B2M | ERAP2 | KRAS | PIM1 | STAT6 |
| BAP1 | ERBB2 | LARP4B | PLCG1 | STK11 |
| BARD1 | ERBB3 | LATS1 | PMS1 | SUFU |
| BCL2 | ERBB4 | LATS2 | PMS2 | TAP1 |
| BCL2L12 | ERCC2 | MAGOH | POLD1 | TAP2 |
| BCL6 | ERCC4 | MAP2K1 | POLE | TBX3 |
| BCOR | ERRFI1 | MAP2K4 | POT1 | TCF7L2 |
| BLM | ESR1 | MAP2K7 | PPM1D | TERT |
| BMPR2 | ETV6 | MAP3K1 | PPP2R1A | TET2 |
| BRAF | EZH2 | MAP3K4 | PPP2R2A | TGFBR2 |
| BRCA1 | FAM135B | MAPK1 | PPP6C | TNFAIP3 |
| BRCA2 | FANCA | MAPK8 | PRDM1 | TNFRSF14 |
| BRIP1 | FANCC | MAX | PRDM9 | TOP1 |
| CARD11 | FANCD2 | MCL1 | PRKACA | TP53 |
| CASP8 | FANCE | MDM2 | PRKAR1A | TP63 |
| CBFB | FANCF | MDM4 | PTCH1 | TPMT |
| CBL | FANCG | MECOM | PTEN | TPP2 |
| CCND1 | FANCI | MEF2B | PTPN11 | TSC1 |
| CCND2 | FANCL | MEN1 | PTPRT | TSC2 |
| CCND3 | FANCM | MET | PXDNL | U2AF1 |
| CCNE1 | FAT1 | MGA | RAC1 | USP8 |
| CD274 | FBXW7 | MITF | RAD50 | USP9X |
| CD276 | FGF19 | MLH1 | RAD51 | VHL |
| CDC73 | FGF23 | MLH3 | RAD51B | WT1 |
| CDH1 | FGF3 | MPL | RAD51C | XPO1 |
| CDH10 | FGF4 | MRE11 | RAD51D | XRCC2 |
| CDK12 | FGF9 | MSH2 | RAD52 | XRCC3 |
| CDK4 | FGFR1 | MSH3 | RAD54L | YAP1 |
| CDK6 | FGFR2 | MSH6 | RAF1 | YES1 |
| CDKN1A | FGFR3 | MTAP | RARA | ZFHX3 |
| CDKN1B | FGFR4 | MTOR | RASA1 | ZMYM3 |
| CDKN2A | FLT3 | MUTYH | RASA2 | ZNF217 |
| CDKN2B | FLT4 | MYC | RB1 | ZNF429 |
| CDKN2C | FOXA1 | MYCL | RBM10 | ZRSR2 |
| CHD4 | FUBP1 | MYCN | RECQL4 | |
| CHEK1 | FYN | MYD88 | RET |
Fusions
| AKT2 | ERG | KRAS | NTRK3 | RB1 |
| ALK | ESR1 | MDM4 | NUTM1 | RELA |
| AR | ETV1 | MET | PDGFRA | RET |
| AXL | ETV4 | MYB | PDGFRB | ROS1 |
| BRAF | ETV5 | MYBL1 | PIK3CA | RSPO2 |
| BRCA1 | FGFR1 | NF1 | PPARG | RSPO3 |
| BRCA2 | FGFR2 | NOTCH1 | PRKACA | TERT |
| CDKN2A | FGFR3 | NOTCH4 | PRKACB | |
| EGFR | FGR | NRG1 | PTEN | |
| ERBB2 | FLT3 | NTRK1 | RAD51B | |
| ERBB4 | JAK2 | NTRK2 | RAF1 |
Full Coding DNA Sequence (including HRR)
Bold and Italics = HRR Genes
|
ABRAXAS1 |
CREBBP | HDAC2 | PARP1 | RUNX1 |
|
ACVR1B |
CSMD3 | HDAC9 | PARP2 | RUNX1T1 |
|
ACVR2A |
CTCF | HLA-A | PARP3 | SDHA |
|
ADAMTS12 |
CTLA4 | HLA-B | PARP4 | SDHB |
|
ADAMTS2 |
CUL3 | HNF1A | PBRM1 | SDHC |
|
AMER1 |
CUL4A | ID3 | PDCD1 | SDHD |
|
APC |
CUL4B | INPP4B | PDCD1LG2 | SETD2 |
|
ARHGAP35 |
CYLD | JAK1 | PDIA3 | SLX4 |
|
ARID1A |
CYP2C9 | JAK2 | PGD | SMAD2 |
|
ARID1B |
CYP2D6 | JAK3 | PHF6 | SMAD4 |
|
ARID2 |
DAXX | KDM5C | PIK3R1 | SMARCA4 |
|
ARID5B |
DDX3X | KDM6A | PMS1 | SMARCB1 |
|
ASXL1 |
DICER1 | KEAP1 | PMS2 | SOCS1 |
|
ASXL2 |
DNMT3A | KLHL13 | POLD1 | SOX9 |
|
ATM |
DOCK3 | KMT2A | POLE | SPEN |
|
ATR |
DPYD | KMT2B | POT1 | STAG2 |
|
ATRX |
DSC1 | KMT2C | PPM1D | STAT1 |
|
AXIN1 |
DSC3 | KMT2D | PPP2R2A | STK11 |
|
AXIN2 |
ELF3 | LARP4B | PRDM1 | SUFU |
|
B2M |
ENO1 | LATS1 | PRDM9 | TAP1 |
|
BAP1 |
EP300 | LATS2 | PRKAR1A | TAP2 |
|
BARD1 |
EPCAM | MAP2K4 | PSMB10 | TBX3 |
|
BCOR |
EPHA2 | MAP2K7 | PSMB8 | TCF7L2 |
|
BLM |
ERAP1 | MAP3K1 | PSMB9 | TET2 |
|
BMPR2 |
ERAP2 | MAP3K4 | PTCH1 | TGFBR2 |
|
BRCA1 |
ERCC2 | MAPK8 | PTEN | TMEM132D |
|
BRCA2 |
ERCC4 | MEN1 | PTPRT | TNFAIP3 |
|
BRIP1 |
ERCC5 | MGA | RAD50 | TNFRSF14 |
|
CALR |
ERRFI1 | MLH1 | RAD51 | TP53 |
|
CASP8 |
ETV6 | MLH3 | RAD51B | TP63 |
|
CBFB |
FANCA | MRE11 | RAD51C | TPP2 |
|
CD274 |
FANCC | MSH2 | RAD51D | TSC1 |
|
CD276 |
FANCD2 | MSH3 | RAD52 | TSC2 |
|
CDC73 |
FANCE | MSH6 | RAD54L | UGT1A1 |
|
CDH1 |
FANCF | MTAP | RASA1 | USP9X |
|
CDH10 |
FANCG | MTUS2 | RASA2 | VHL |
|
CDK12 |
FANCI | MUTYH | RB1 | WT1 |
|
CDKN1A |
FANCL | NBN | RBM10 | XRCC2 |
|
CDKN1B |
FANCM | NCOR1 | RECQL4 | XRCC3 |
|
CDKN2A |
FAS | NF1 | RNASEH2A | ZBTB20 |
|
CDKN2B |
FAT1 | NF2 | RNASEH2B | ZFHX3 |
|
CDKN2C |
FBXW7 | NOTCH1 | RNASEH2C | ZMYM3 |
|
CHEK1 |
FUBP1 | NOTCH2 | RNF43 | ZRSR2 |
|
CHEK2 |
GATA3 | NOTCH3 | RPA1 | |
|
CIC |
GNA13 | NOTCH4 | RPL22 | |
|
CIITA |
GPS2 | PALB2 | RPL5 |
TEST ORDERING
The intended use of the AuraSeq-Comprehensive test is to detect somatic variants in multiple cancer driver genes and genomic signatures using NGS technology in advanced carcinoma samples to help guide therapeutic decisions, especially in those patients who have failed to respond to conventional chemotherapy or are about to start a TKI regime, Immune Checkpoint Inhibition (ICI), or PARP inhibitors (PARPi) therapies.
Physicians (or other individuals authorized by law to order tests) should only order tests that are medically necessary for the diagnosis or treatment of the patient.
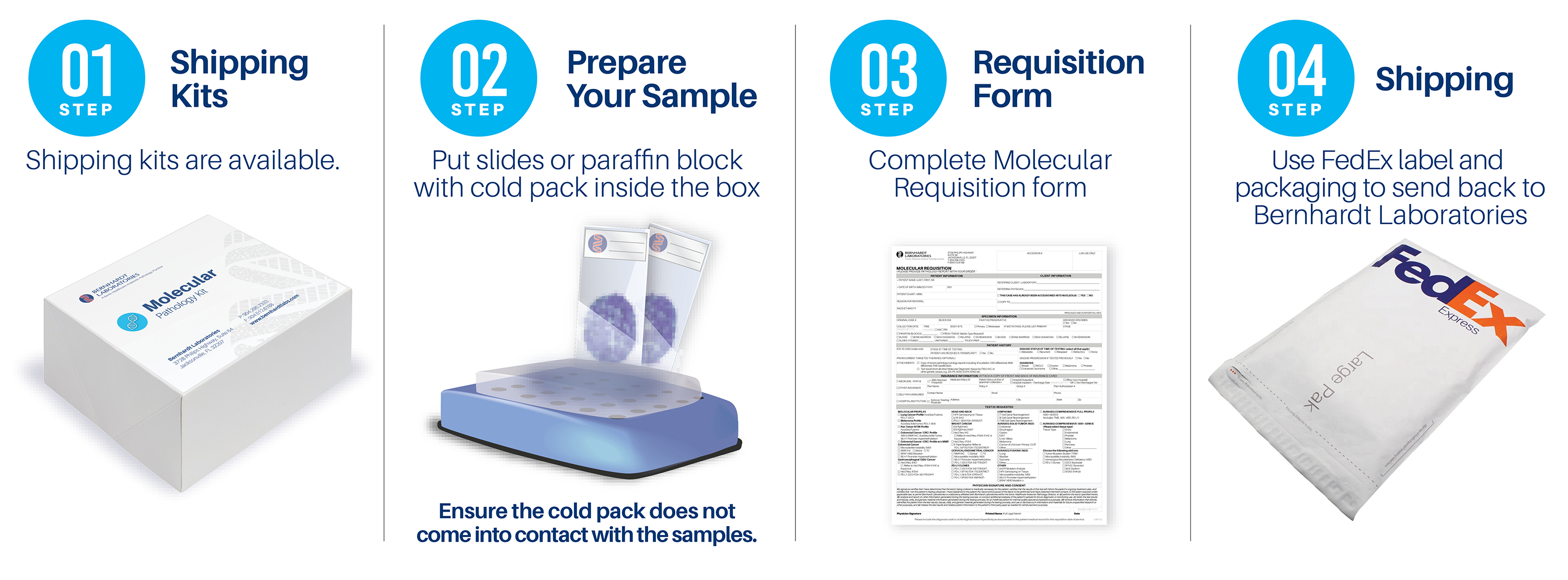
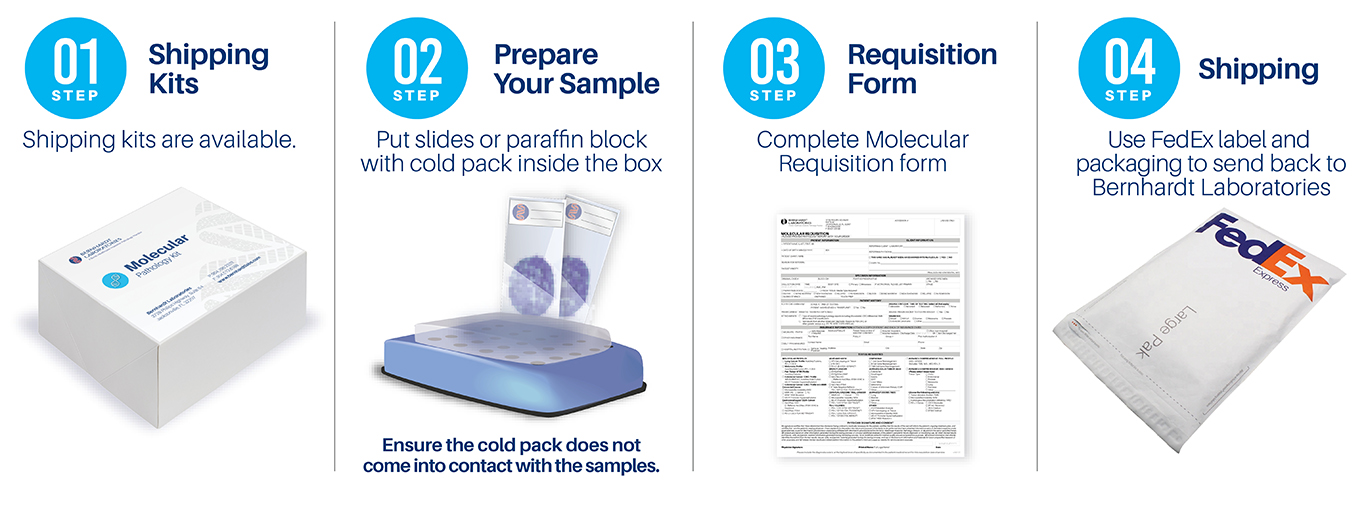
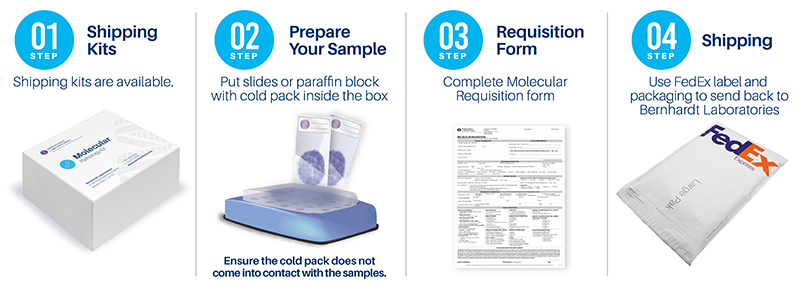
Request shipping kits or requisitions:
or 904.296.2333
Ship materials to:
Bernhardt Laboratories
Attn: Molecular Department
3728 Philips Highway, Suite 64
Jacksonville, FL 32207
Specimen Requirements
- Compatible with formalin-fixed, paraffin-embedded (FFPE) tissue samples
- 1 H&E + 4-5 unstained/unbaked slides
- Compatible with cytology smears (1-2 slides)
- Requires minimal nucleic acid input (2 to 20 ng total of DNA and/or RNA)
References:
1 Data on ~2300 samples submitted for AuraSeq testing.
2 Nature. 2018;553(7689):446-454.
3 JAMA. 2021;325(7):669-685.
4 Cancer Cytopathol. 2019;127(5):285-296.
5 J Clin Pathol. 2021;207825.
6 Data on ~2300 samples submitted for AuraSeq testing.
7 J Oncol Pract. 2016;12(4):e396-404.
This test was developed and its performance characteristics determined by Sonic Healthcare USA, Anatomic Pathology and Bernhardt Laboratories. It has not been cleared or approved by the U.S. Food and Drug Administration (FDA). The FDA has determined that such clearance or approval is not necessary. This test is used for clinical purposes and should not be regarded as investigational or for research. Bernhardt Laboratories is qualified to perform high complexity testing under the Clinical Laboratory Improvement Amendments (CLIA). This test is considered an LDT- Laboratory Developed Test.
Copyright © 2022 Sonic Healthcare USA, Inc. All rights reserved. All of the information in this document is the property of Sonic Healthcare USA. It may not be distributed, transmitted, reproduced, copied or displayed without the written permission of Sonic Healthcare USA. Sonic Healthcare USA, including its affiliates, does not dispense medical advice. The content in this marketing collateral is intended for informational purposes only and does not constitute legal, medical or any professional advice.
Auraseq Comprehensive samples can not be recieved by the states of New York or California.